Gene Expression Profile Prediction in Uveal Melanoma Using Deep Learning: A Pilot Study for the Development of an Alternative Survival Prediction Tool
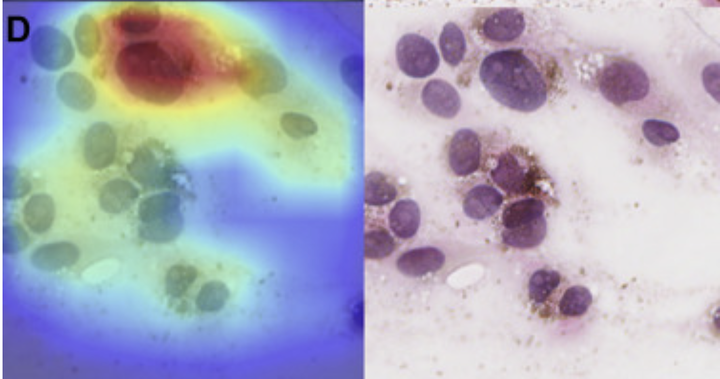 Sample CAM maps.
Sample CAM maps.
Abstract
In recent years, artificial intelligence, especially deep learning (DL), has generated immense interest in the medical field. Deep learning has been used to classify medical images in disciplines such as ophthalmology and oncologic pathology. One commonality across malignancies is that cancer cell morphologic features potentially reflect the underlying genetics and that careful analysis of cytopathologic characteristics often provides helpful prognostication information. However, detailed measurement and analysis of cell morphologic features are labor intensive and clinically infeasible, and thus is limited largely to research. Analyses of pathologic images to extract useful information ultimately are a pattern recognition exercise in which DL excels. We hypothesize that DL methods, when applied appropriately in cytopathologic image analysis, could predict patient outcomes that correlate with the tumors’ genetic or molecular profiles, or both. Our disease of interest is uveal melanoma (UM), which is unique among malignancies for having a validated prognostic gene expression profile (GEP) test that can be used independently of other clinicopathologic parameters and can be tested on fine-needle aspiration biopsy (FNAB) samples. Patients with UM can be divided into 2 classes by GEP, with a survival probability of 95% in class 1 patients and 31% in class 2 patients at 92 months.1,2 Our ultimate goal is to develop a DL-based image analytic tool for survival prognostication in UM. Given that GEP is correlated highly with survival in UM, we set out to conduct a pilot study to develop a DL system that can distinguish patient survival using smeared cytologic aspirates from FNAB samples and GEP as the reference standard.