Biography
I am a Machine Learning Engineer in ByteDance working in machine auditing for TikTok Shop products. I graduated as a Computer Science Ph.D. from Johns Hopkins University with a background in interpretable computer vision systems for medical image analysis with human-computer interaction, image classification, object detection, and segmentation. I have rich experience with whole slide images, CT scans, and X-rays. I am the first author of Nature partner journal paper. I have excellent communication skills and ability to work on multi-disciplinary teams.
Download my resumé.
- Computer Vision
- Medical Imaging
- Transparent System
- Human-Computer Intraction
- Generative AI
-
PhD in Computer Science, 2018-2022
Johns Hopkins University
-
M.A. in Statistics, 2016-2017
Columbia University
-
BSc in Physics, 2012-2016
Fudan Univerisity
Skills
Experience
Interpretable Video Translation by generative AI:
- Created the largest dataset of talking head videos from YouTube.
- Established multi-person & lingual audio/video synchronization.
- Refined the facial landmark generation network for better articulation.
- Used diffusion to achieve immersive lip synchronization in videos with translated audio.
2D-3D style transfer for VR:
- Achieved real-time inference.
- Enabled human interaction for personalized customization.
- Preserved 3D visual reality.
- Utilized PyTorch3D & nvdiffrast as differential rendering.
- Preserved object style consistency by semantic style transfer.
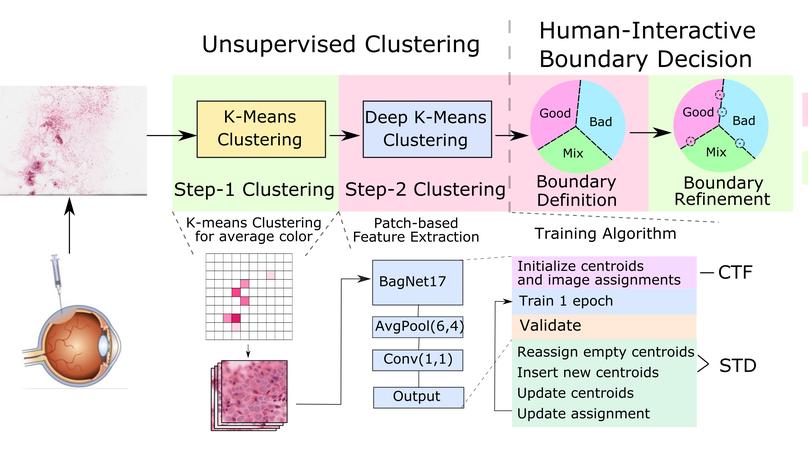
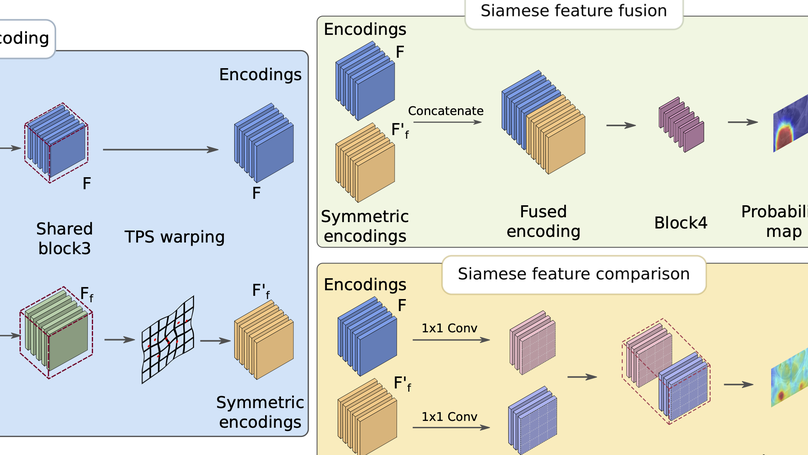
Symmetric learning for Fracture Detection in Pelvic Trauma X-ray:
- Deployed in Chang Gung Memorial Hospital in Taiwan ans used by > 5000 patients.
- Paper accepted by ECCV 2020 with poster.
- Improved AUC from 0.95 to 0.98 and fracture recall from 0.89 to 0.93 (FPR=0.1).
- Mimicked radiologists to detect fractures by comparing bilateral symmetric regions.
- Focused on anatomical asymmetry with contrastive learning.
Deep Hierarchical Multi-label Classification of Chest X-ray:
- Paper accepted by MIDL 2019 with oral presentation.
- Paper accepted by Journal “Medical Image Analysis”.
- Mimicked radiologists to classify abnormality with clinical taxonomy.
- Improved classification AUC from 0.87 to 0.89.
- Robust to incompletely labeled data and preserved 85% performance drop.
Lung nodule detection in CT images:
- Achieved rank 6/2887 teams in the Skylake competition by Intel and Alibaba.
- Applied PyTorch, 3D UNet and Caffe, Faster RCNN in 1000 CT scans.
- Used fusion method to achieve false positive reduction.
Featured Publications
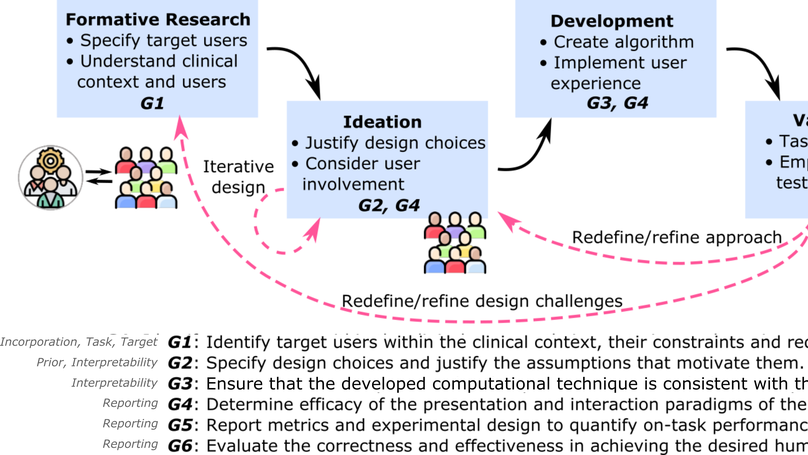
The Need for a More Human-Centered Approach to Designing and Validating Transparent AI in Medical Image Analysis - Guidelines and Evidence from a Systematic Review.
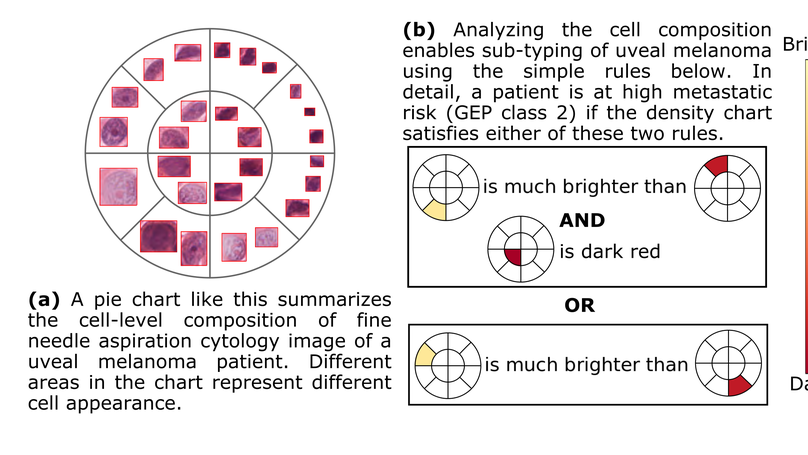
An interpretable classification of genetic information for UM prognostication with cell composition analysis.
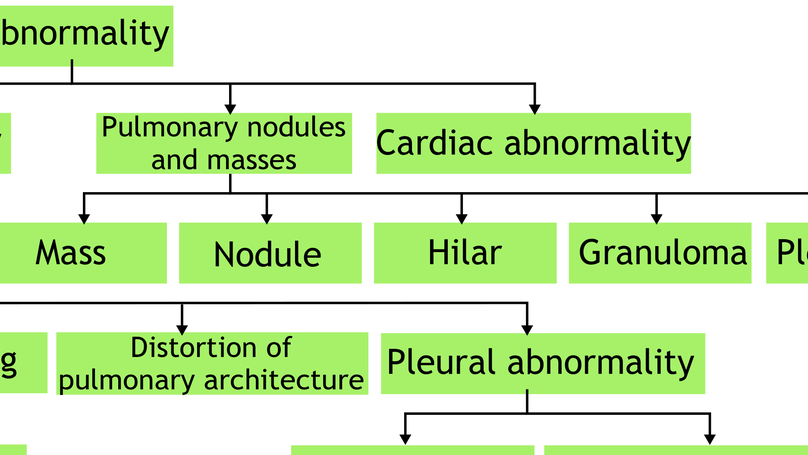
A two-stage hierarchical multi-label classification algorithm for chest X-ray abnormality classification.